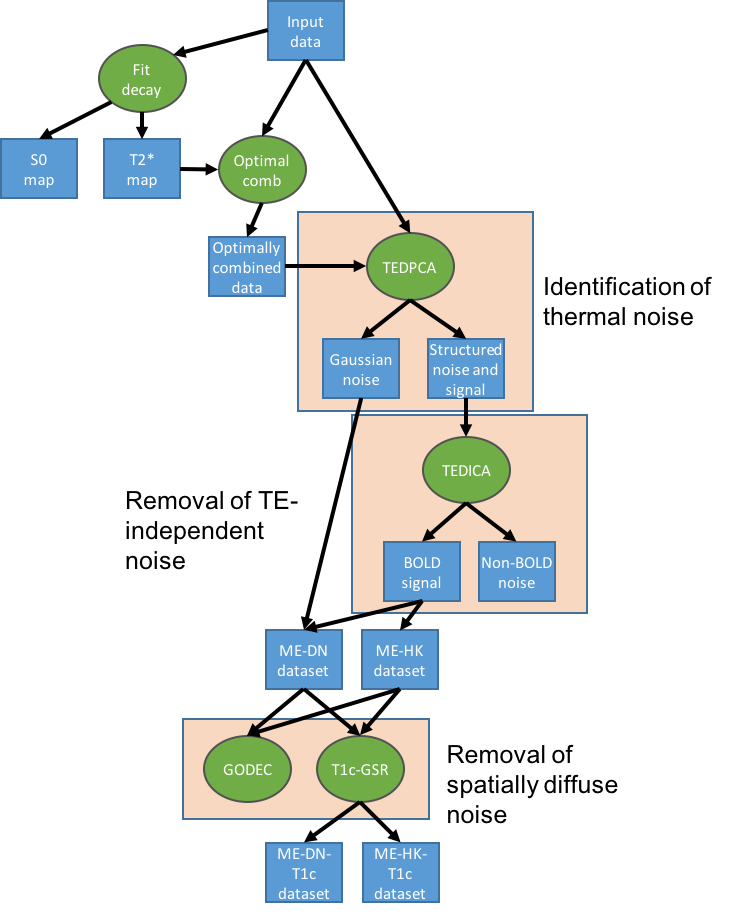
Processing pipeline details¶
tedana works by decomposing multi-echo BOLD data via PCA and ICA.
These components are then analyzed to determine whether they are TE-dependent
or -independent. TE-dependent components are classified as BOLD, while
TE-independent components are classified as non-BOLD, and are discarded as part
of data cleaning.
In tedana, we take the time series from all the collected TEs, combine them,
and decompose the resulting data into components that can be classified as BOLD
or non-BOLD. This is performed in a series of steps, including:
- Principal components analysis
- Independent components analysis
- Component classification
Multi-echo data¶
Here are the echo-specific time series for a single voxel in an example resting-state scan with 5 echoes.
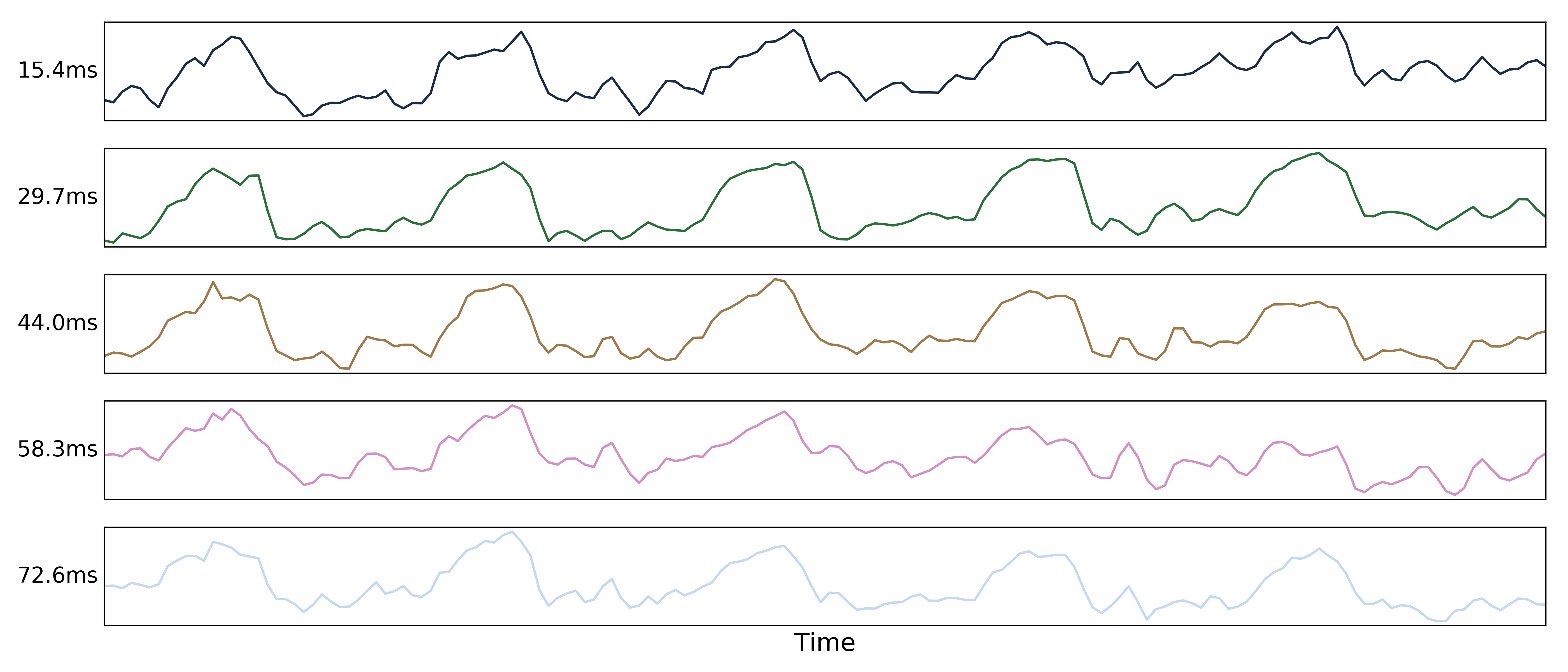
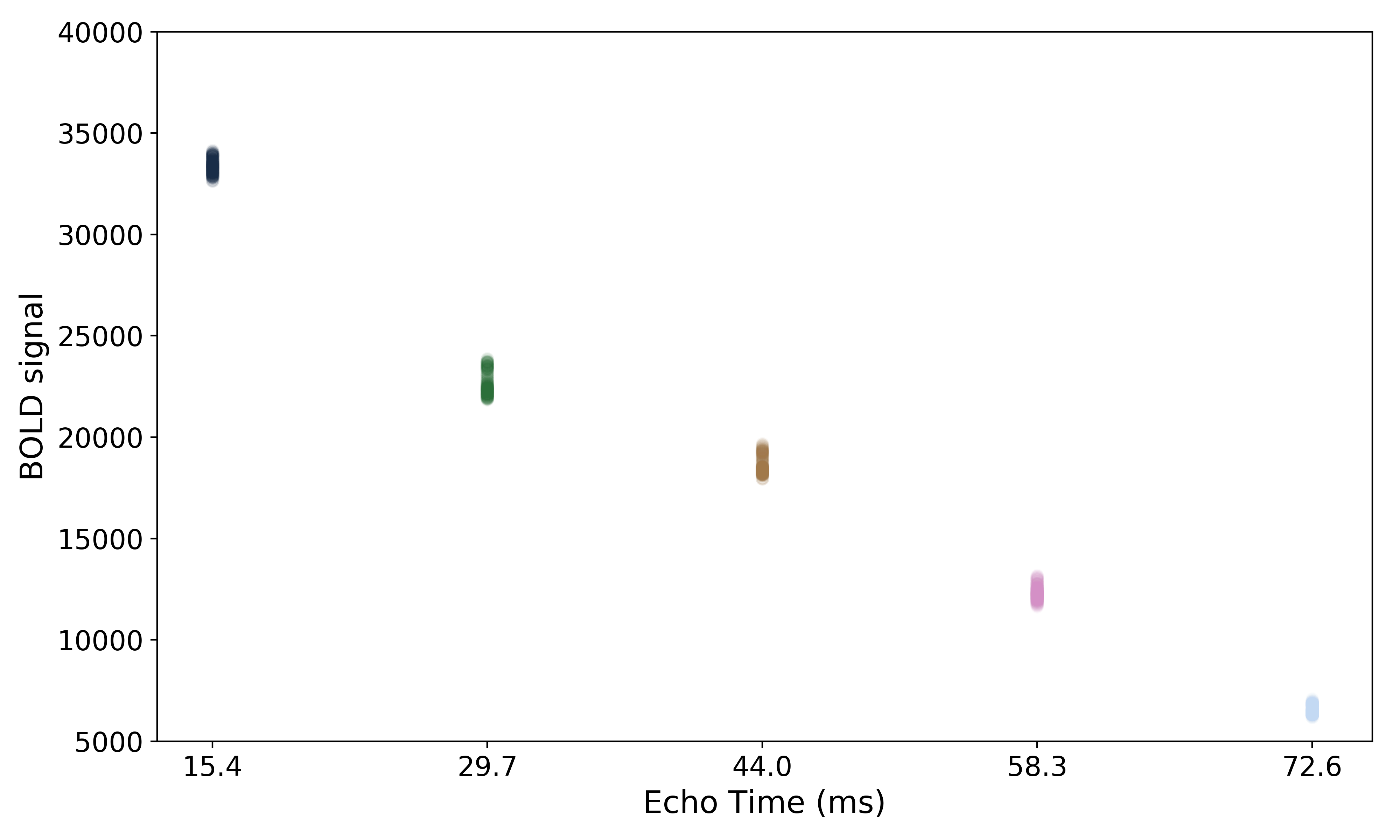
The values across volumes for this voxel scale with echo time in a predictable manner.
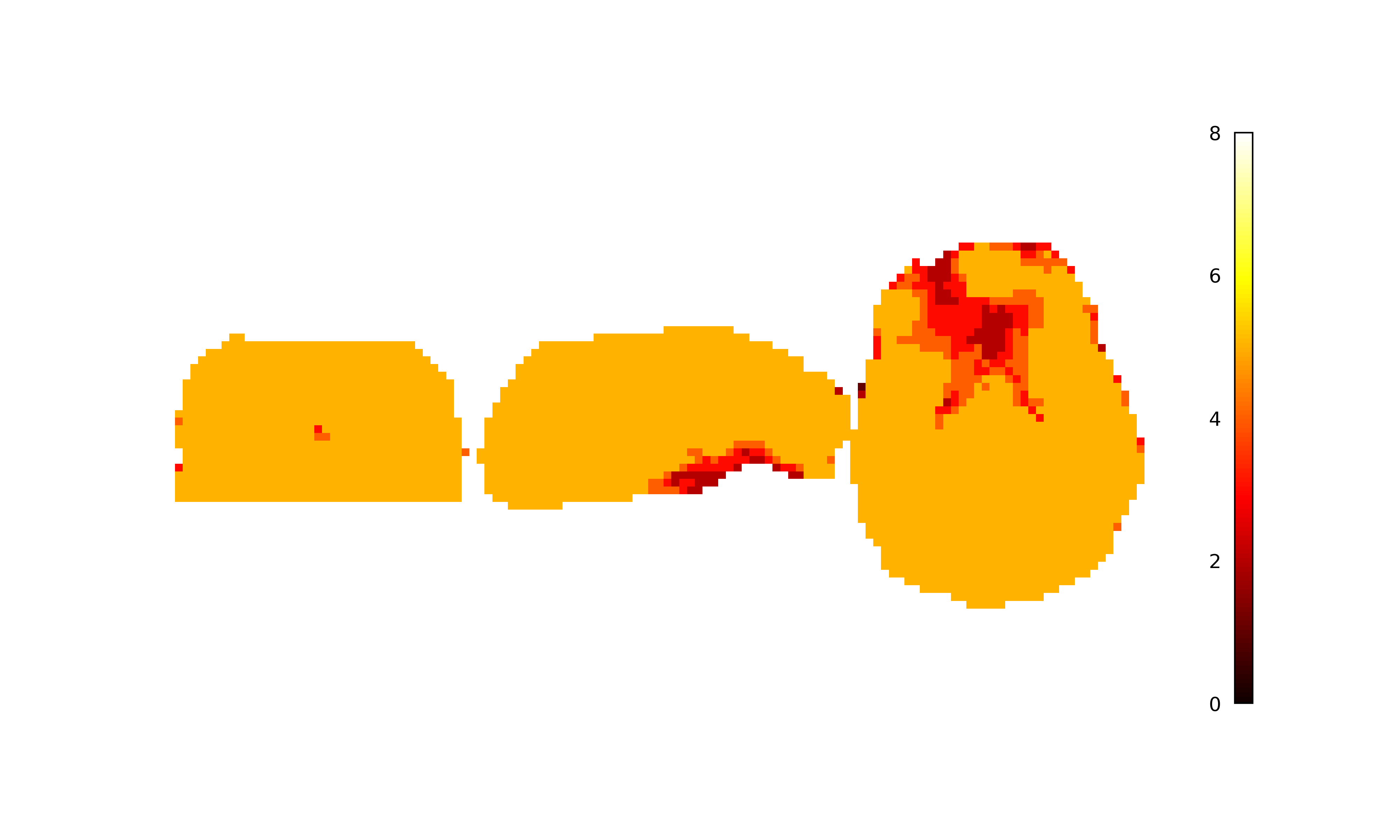
Adaptive mask generation¶
Longer echo times are more susceptible to signal dropout, which means that
certain brain regions (e.g., orbitofrontal cortex, temporal poles) will only
have good signal for some echoes. In order to avoid using bad signal from
affected echoes in calculating 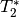 and
and 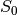 for a given voxel,
for a given voxel,
tedana generates an adaptive mask, where the value for each voxel is the
number of echoes with “good” signal. When 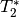 and
and 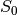 are
calculated below, each voxel’s values are only calculated from the first
are
calculated below, each voxel’s values are only calculated from the first 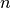 echoes, where
echoes, where 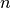 is the value for that voxel in the adaptive mask.
is the value for that voxel in the adaptive mask.
Monoexponential decay model fit¶
The next step is to fit a monoexponential decay model to the data in order to
estimate voxel-wise 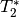 and
and 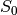 .
.
In order to make it easier to fit the decay model to the data, tedana
transforms the data. The BOLD data are transformed as 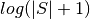 , where
, where
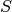 is the BOLD signal. The echo times are also multiplied by -1.
is the BOLD signal. The echo times are also multiplied by -1.
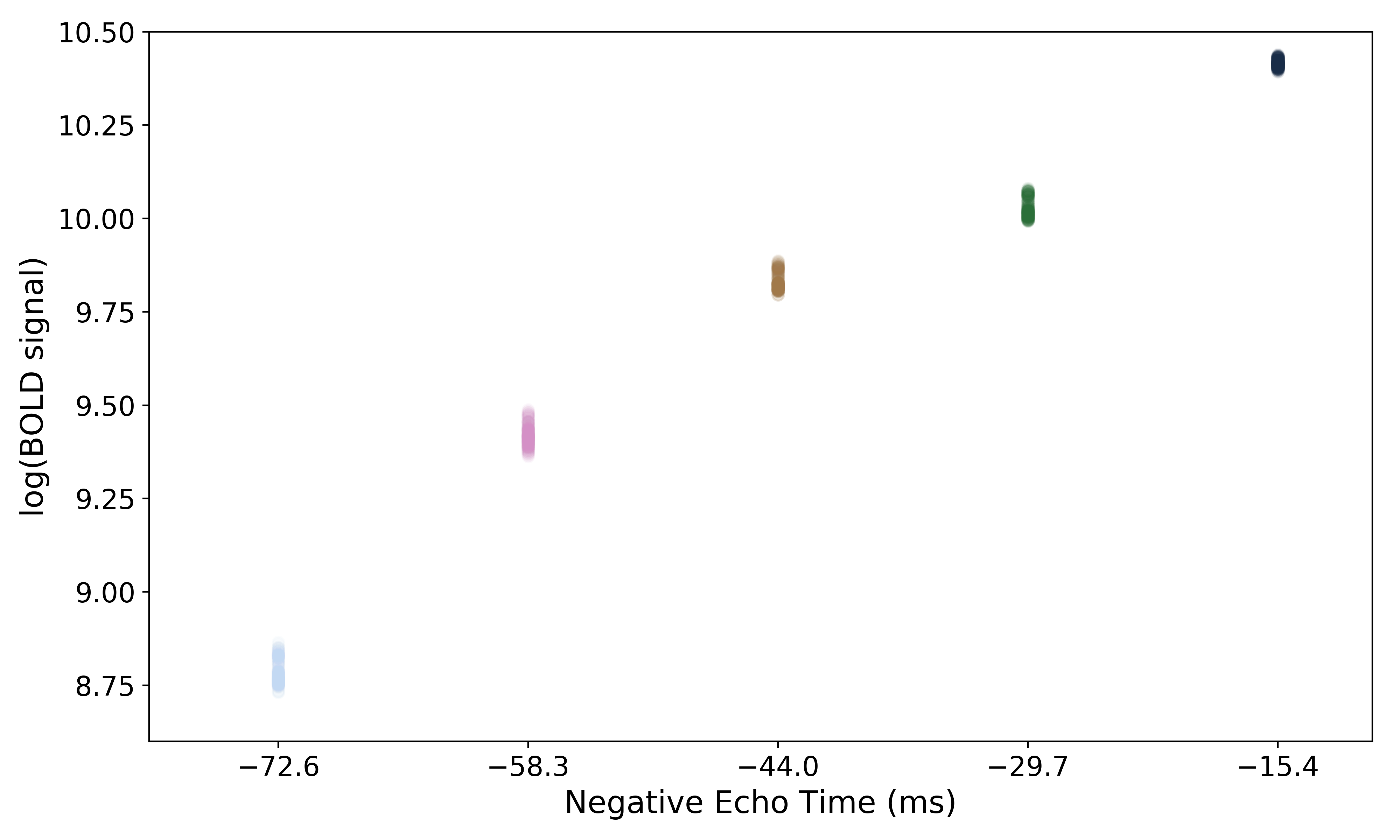
A simple line can then be fit to the transformed data with linear regression. For the sake of this introduction, we can assume that the example voxel has good signal in all five echoes (i.e., the adaptive mask has a value of 5 at this voxel), so the line is fit to all available data.
Note
tedana actually performs and uses two sets of 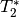 /
/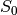 model fits.
In one case,
model fits.
In one case, tedana estimates 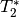 and
and 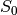 for voxels with good signal in at
least two echoes. The resulting “limited”
for voxels with good signal in at
least two echoes. The resulting “limited” 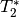 and
and 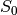 maps are used throughout
most of the pipeline. In the other case,
maps are used throughout
most of the pipeline. In the other case, tedana estimates 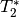 and
and 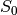 for voxels
with good data in only one echo as well, but uses the first two echoes for
those voxels. The resulting “full”
for voxels
with good data in only one echo as well, but uses the first two echoes for
those voxels. The resulting “full” 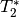 and
and 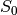 maps are used to generate the
optimally combined data.
maps are used to generate the
optimally combined data.
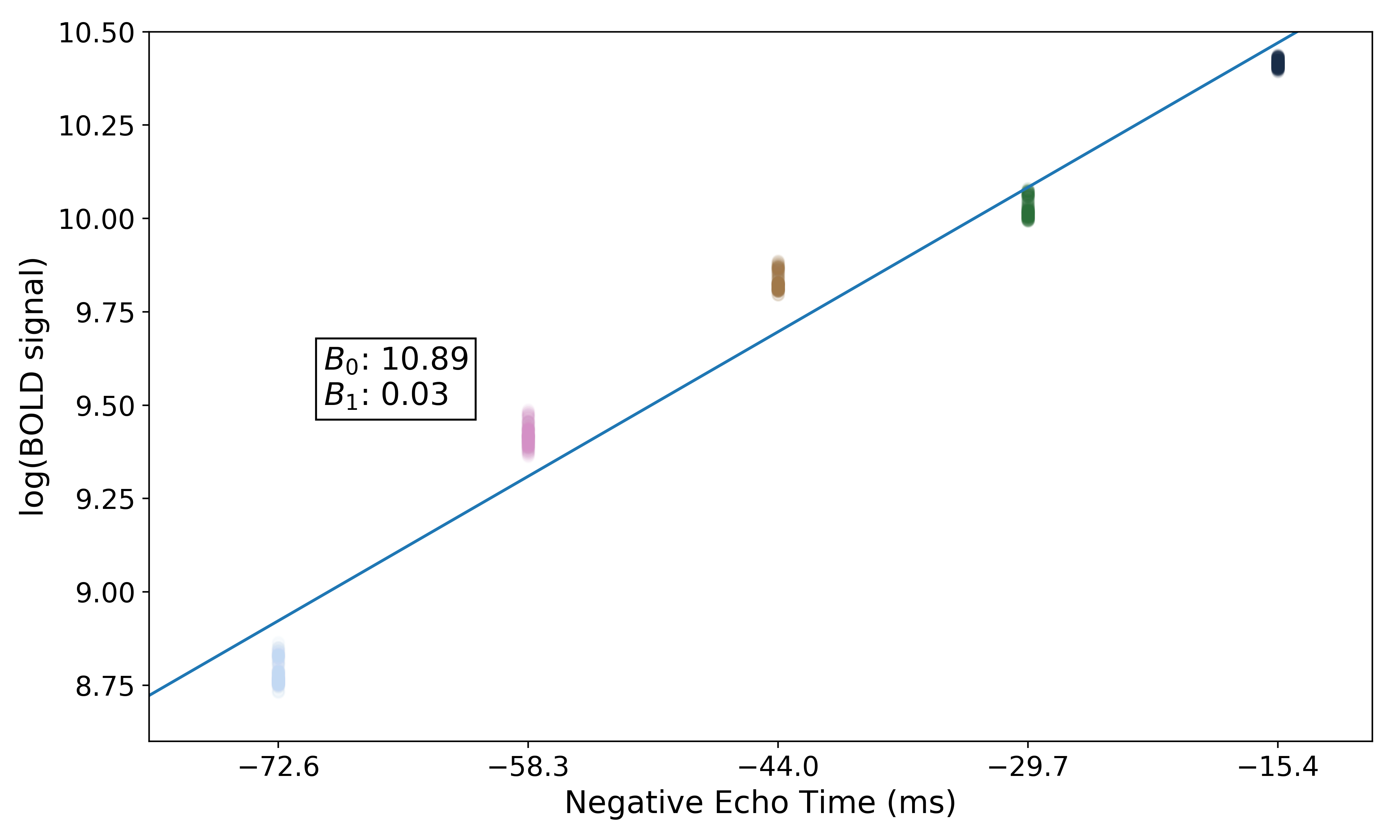
The values of interest for the decay model, 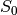 and
and 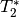 ,
are then simple transformations of the line’s intercept (
,
are then simple transformations of the line’s intercept (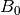 ) and
slope (
) and
slope (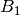 ), respectively:
), respectively:
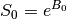
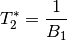
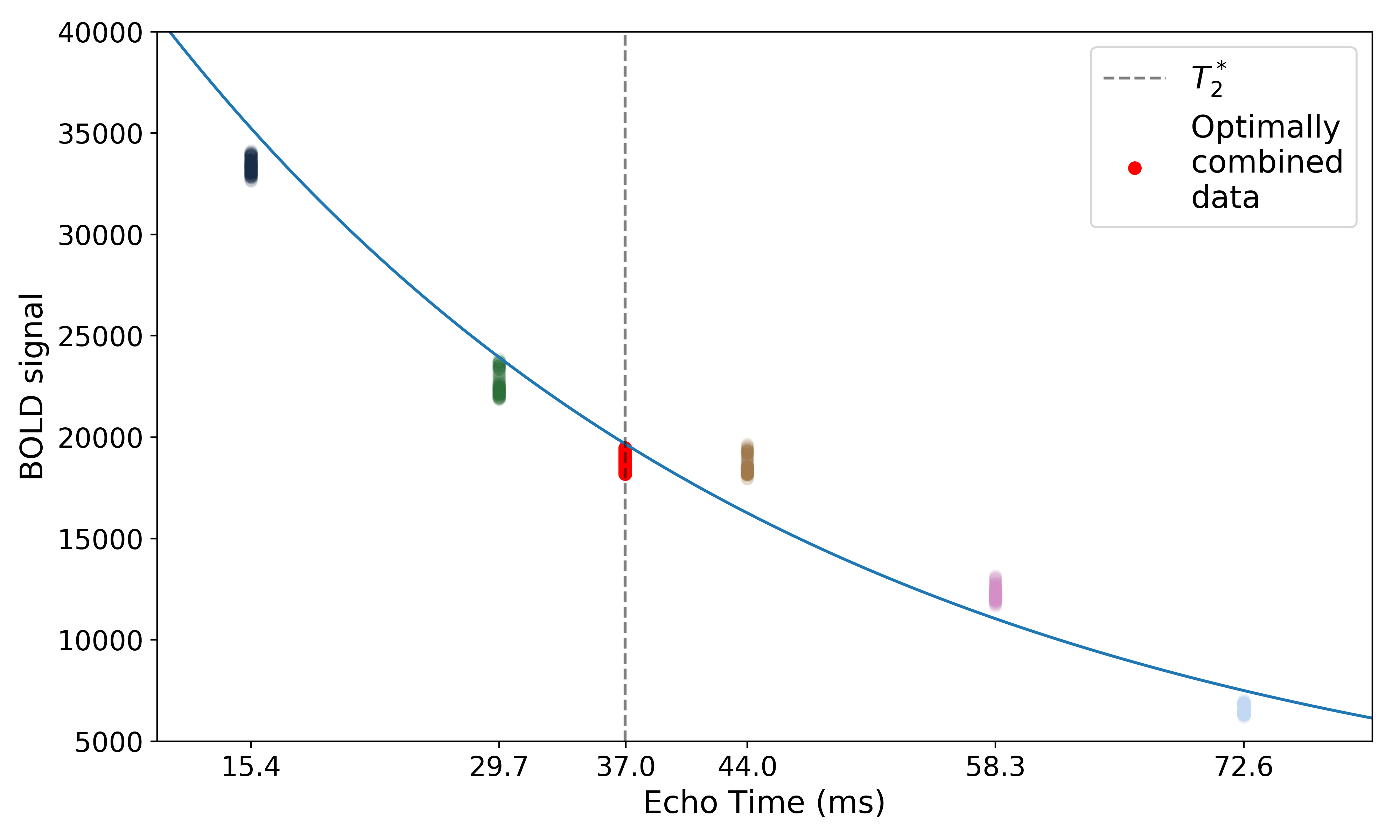
The resulting values can be used to show the fitted monoexponential decay model on the original data.
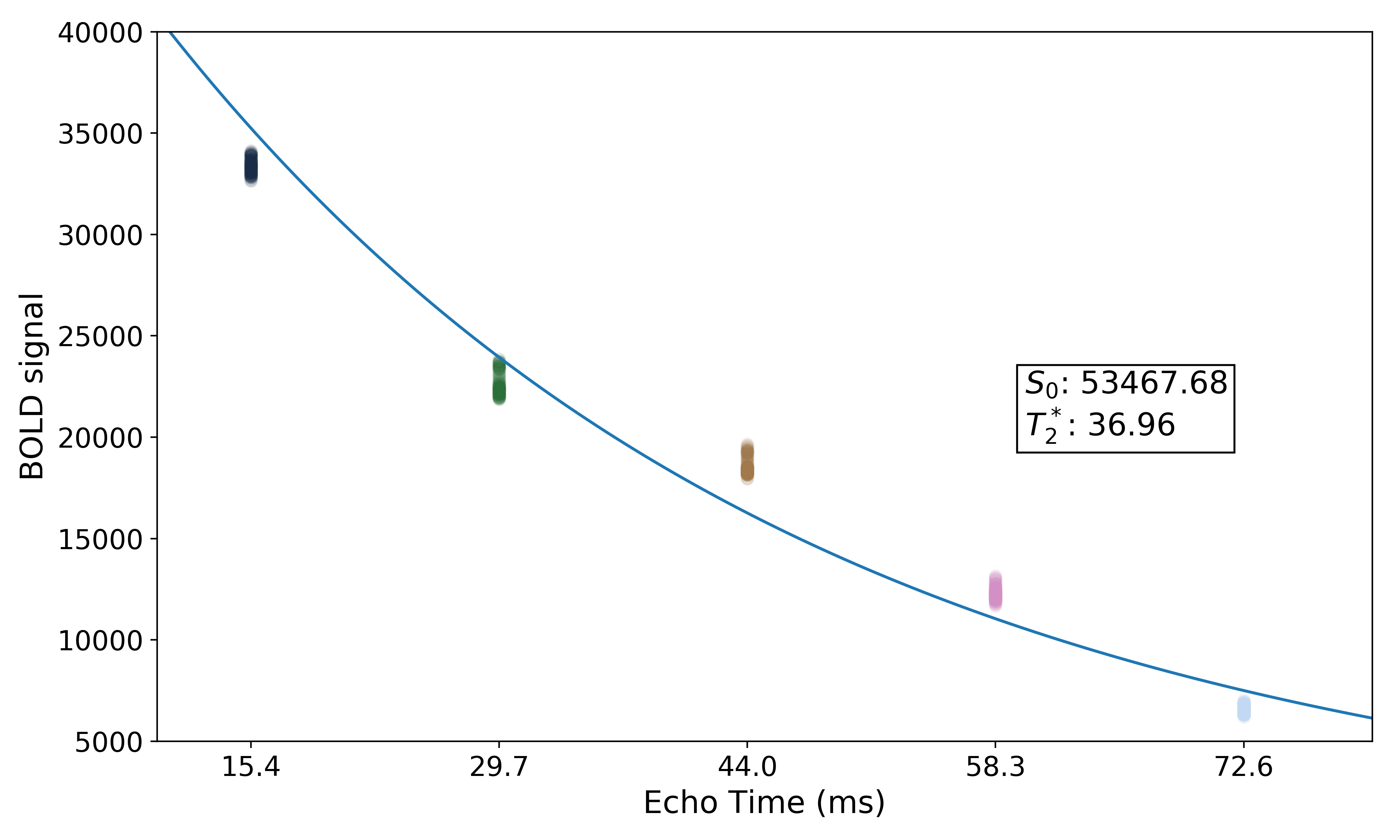
We can also see where 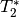 lands on this curve.
lands on this curve.
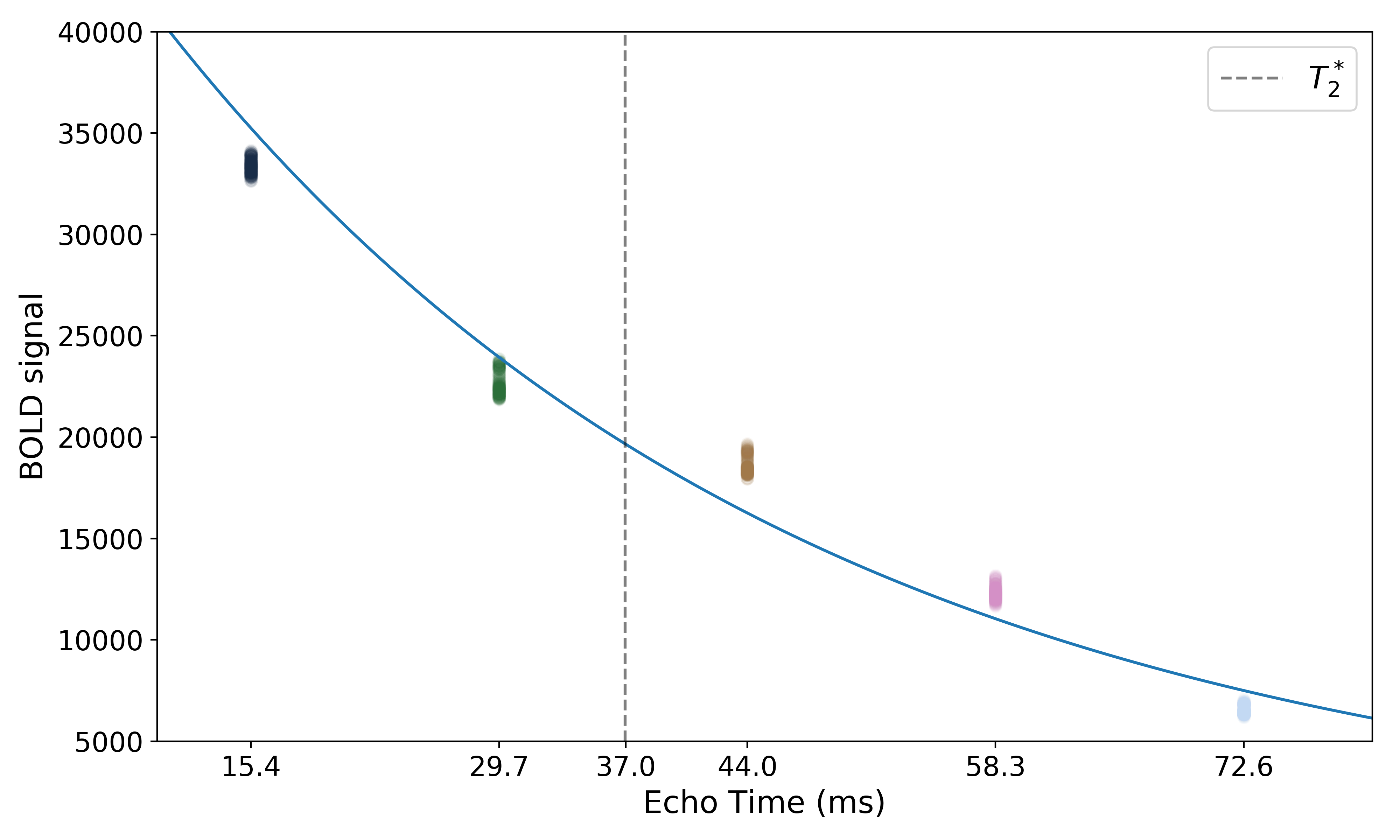
Optimal combination¶
Using the 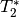 estimates,
estimates, tedana combines signal across echoes using a
weighted average. The echoes are weighted according to the formula
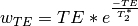
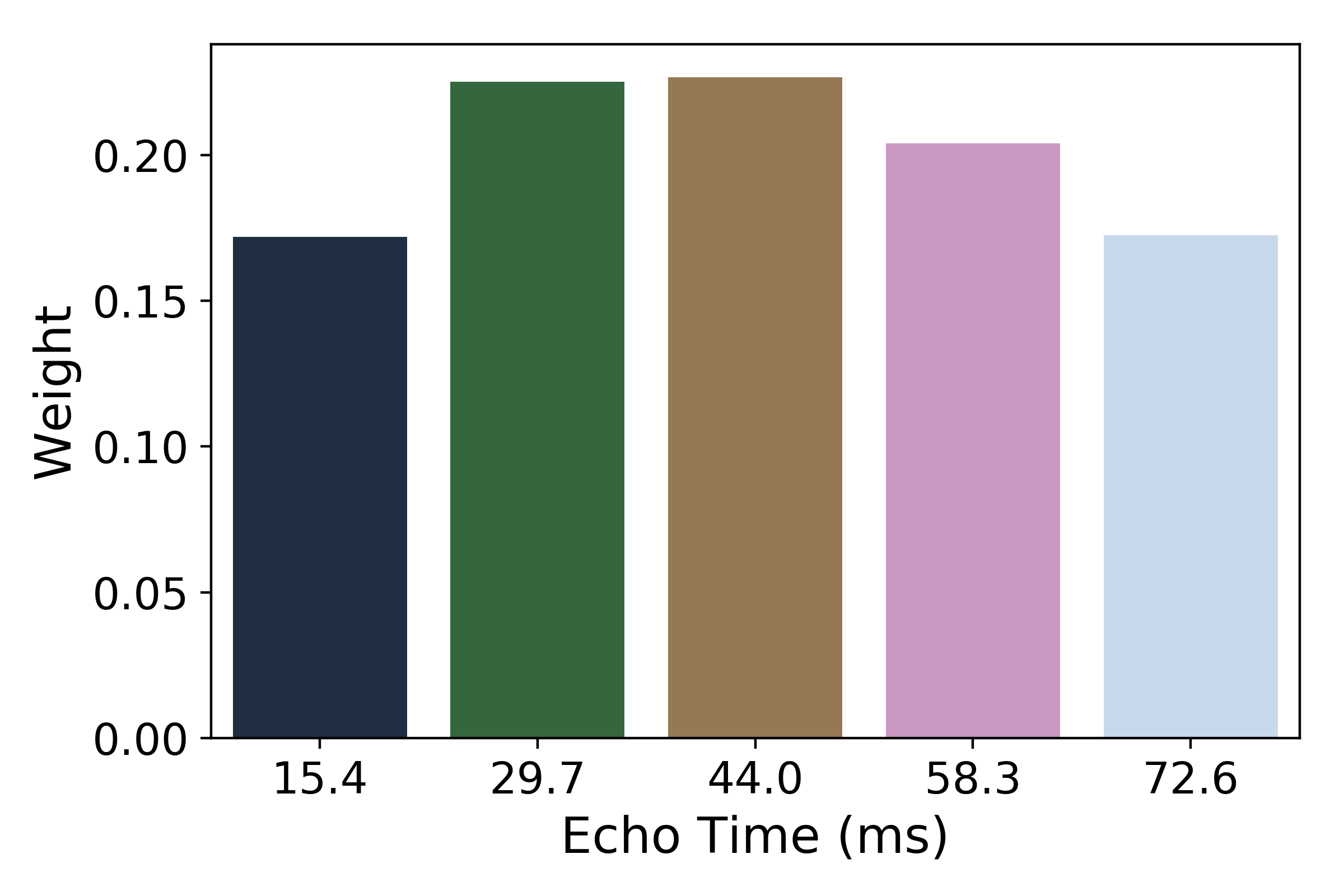
The weights are then normalized across echoes. For the example voxel, the resulting weights are:
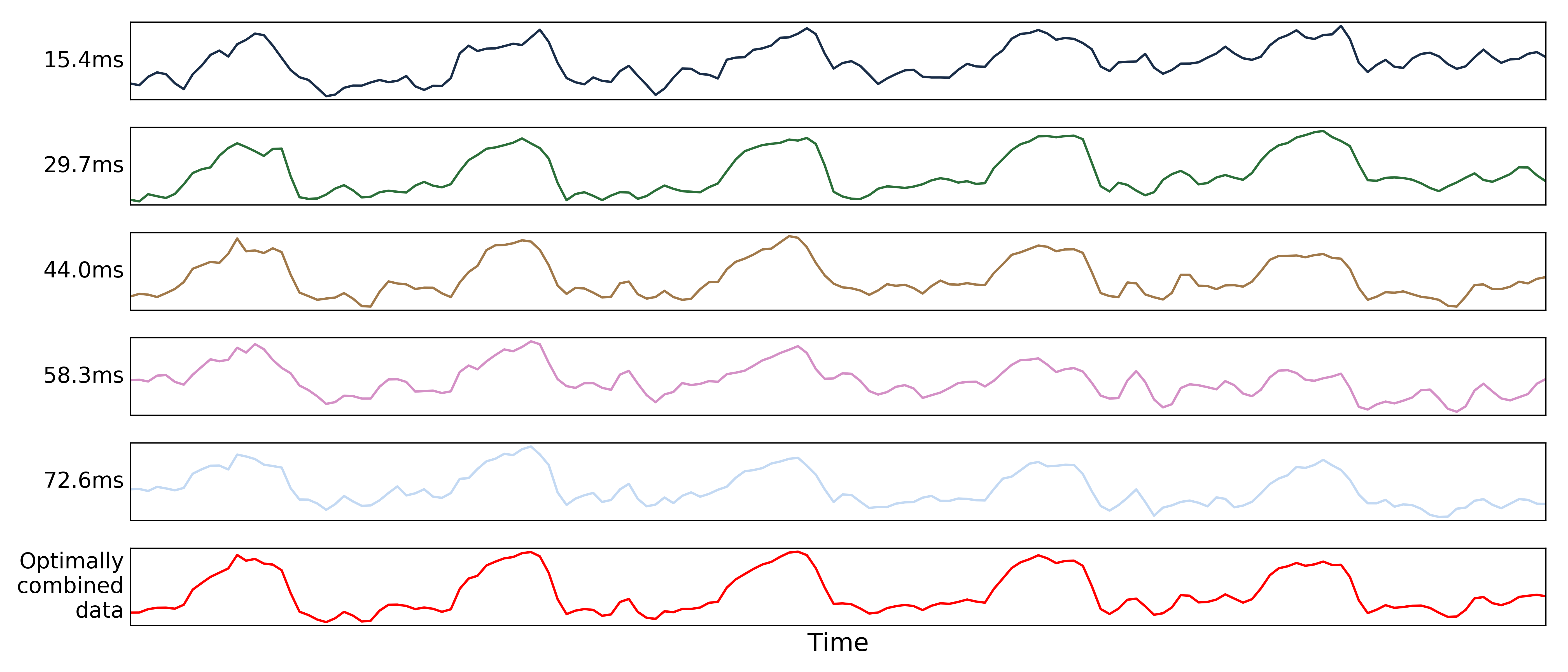
The distribution of values for the optimally combined data lands somewhere between the distributions for other echoes.
The time series for the optimally combined data also looks like a combination of the other echoes (which it is).
TEDPCA¶
The next step is to identify and temporarily remove Gaussian (thermal) noise with TE-dependent principal components analysis (PCA). TEDPCA applies PCA to the optimally combined data in order to decompose it into component maps and time series. Here we can see time series for some example components (we don’t really care about the maps):
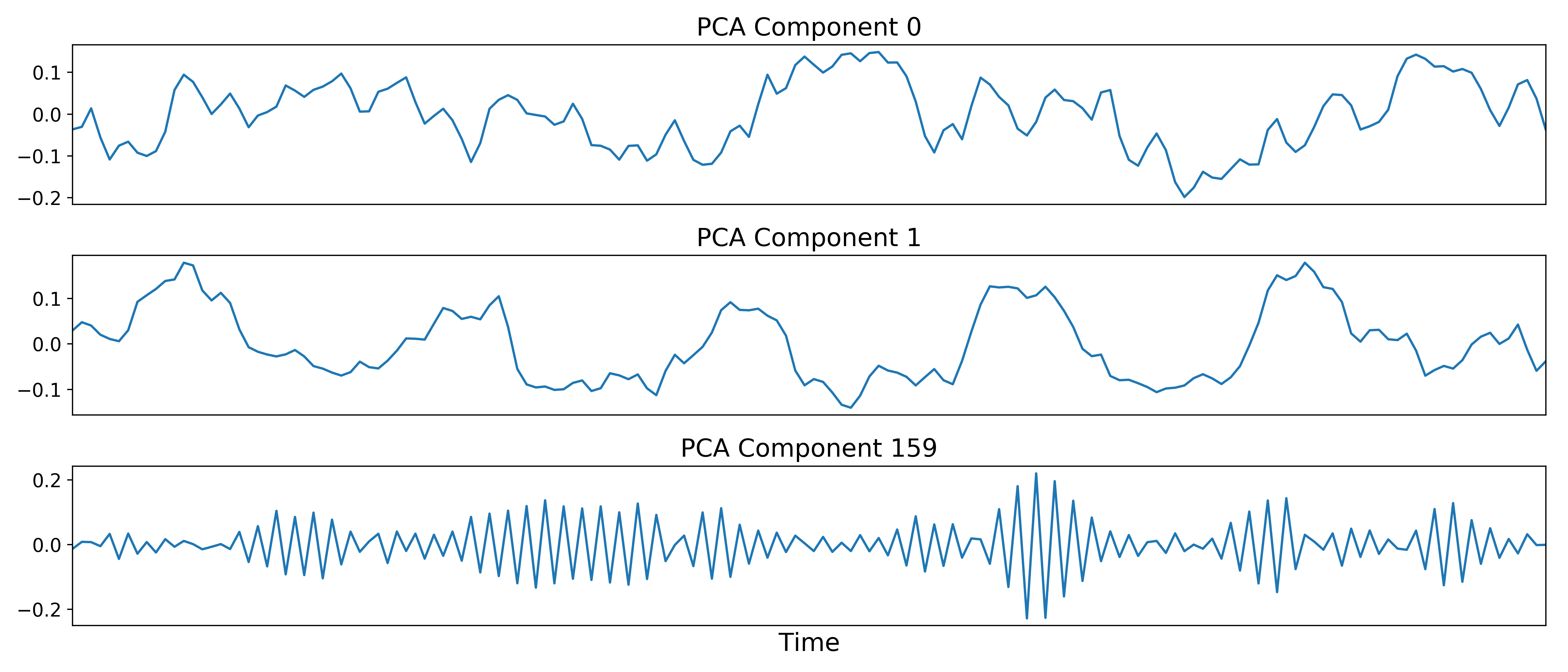
These components are subjected to component selection, the specifics of which vary according to algorithm.
In the simplest approach, tedana uses Minka’s MLE to estimate the
dimensionality of the data, which disregards low-variance components.
A more complicated approach involves applying a decision tree to identify and discard PCA components which, in addition to not explaining much variance, are also not significantly TE-dependent (i.e., have low Kappa) or TE-independent (i.e., have low Rho).
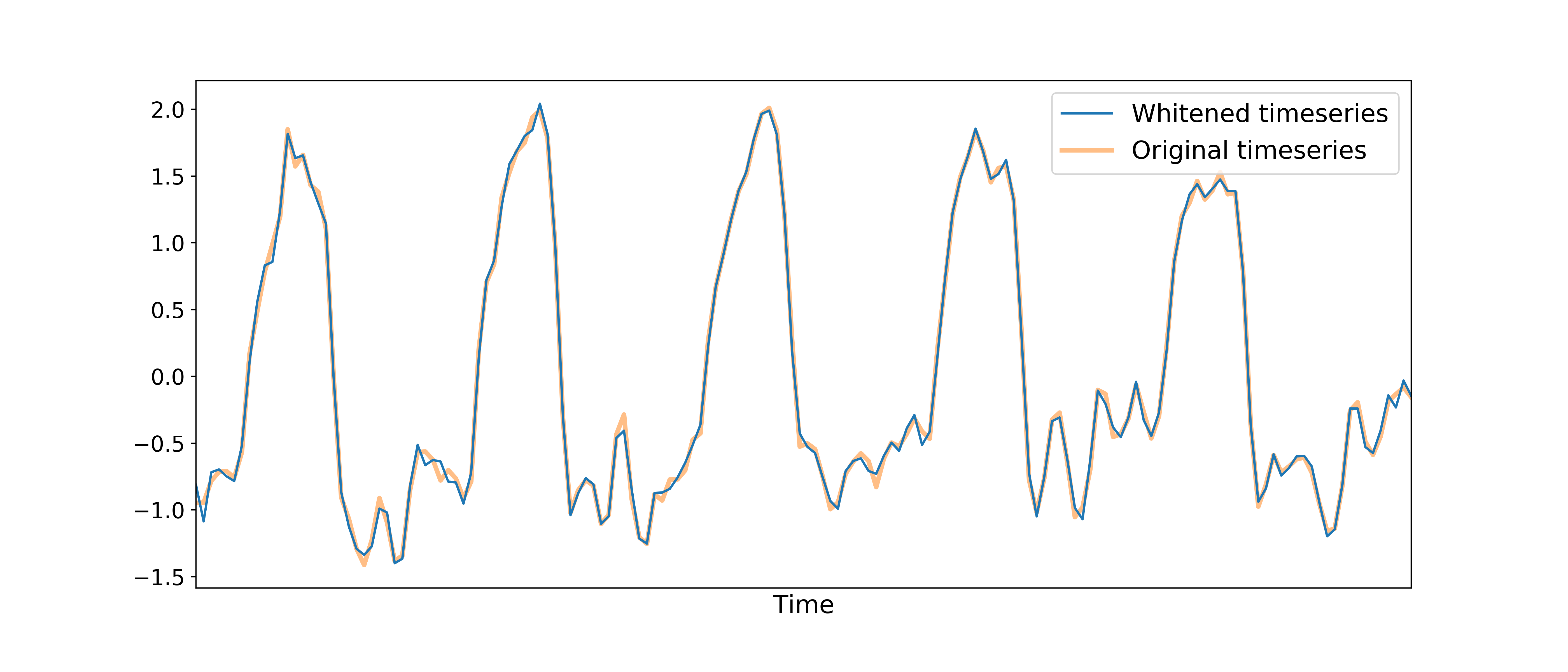
After component selection is performed, the retained components and their associated betas are used to reconstruct the optimally combined data, resulting in a dimensionally reduced (i.e., whitened) version of the dataset.
TEDICA¶
Next, tedana applies TE-dependent independent components analysis (ICA) in
order to identify and remove TE-independent (i.e., non-BOLD noise) components.
The dimensionally reduced optimally combined data are first subjected to ICA in
order to fit a mixing matrix to the whitened data.
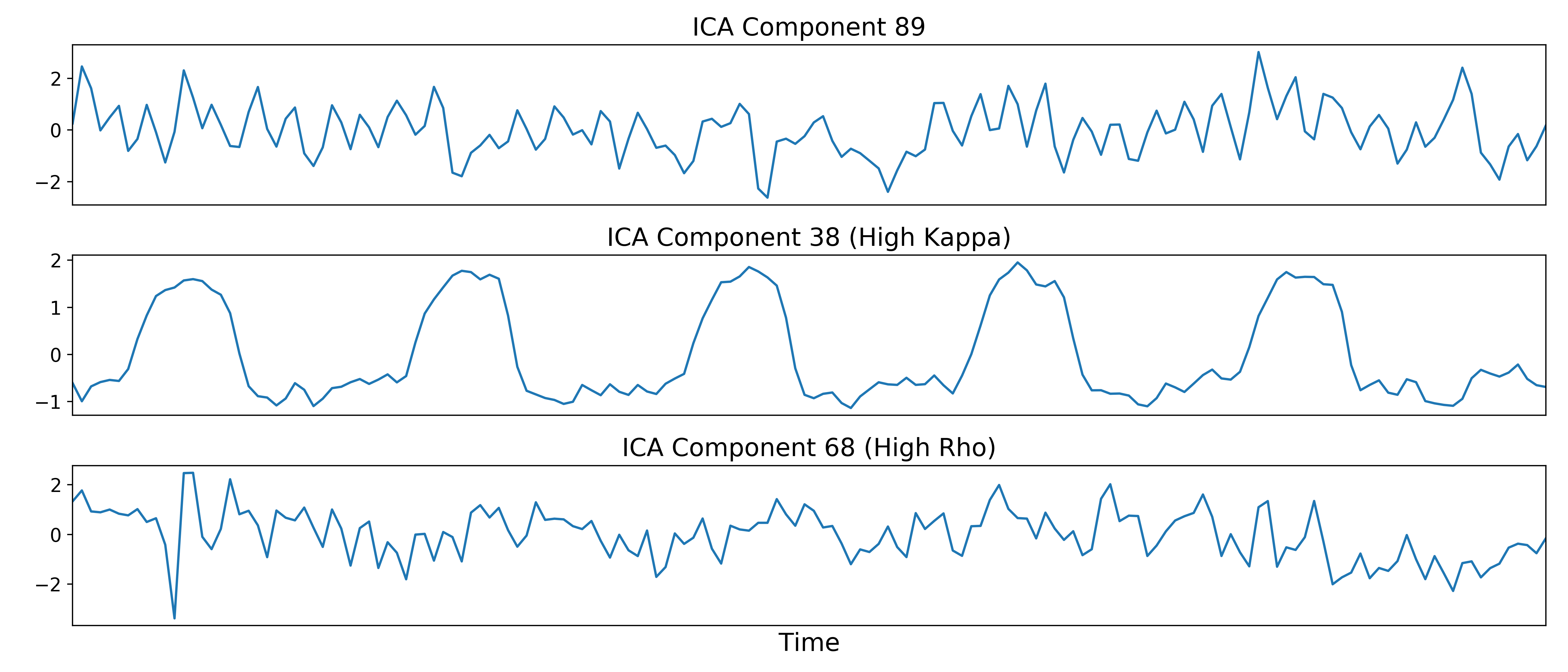
Linear regression is used to fit the component time series to each voxel in each echo from the original, echo-specific data. This way, the thermal noise is retained in the data, but is ignored by the TEDICA process. This results in echo- and voxel-specific betas for each of the components.
TE-dependence (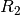 ) and TE-independence (
) and TE-independence (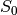 ) models can then
be fit to these betas. These models allow calculation of F-statistics for the
) models can then
be fit to these betas. These models allow calculation of F-statistics for the
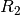 and
and 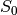 models (referred to as
models (referred to as  and
and
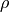 , respectively).
, respectively).
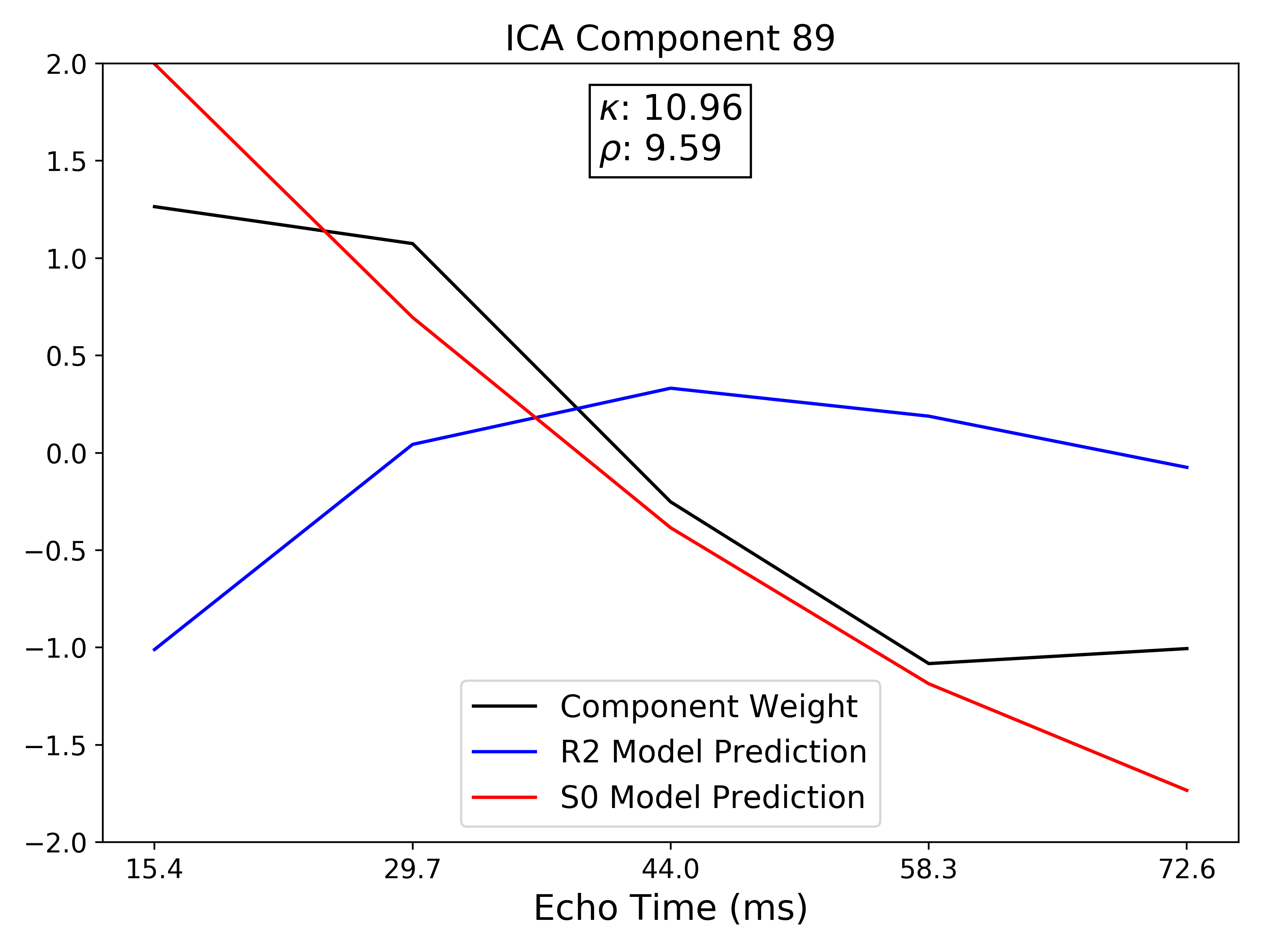
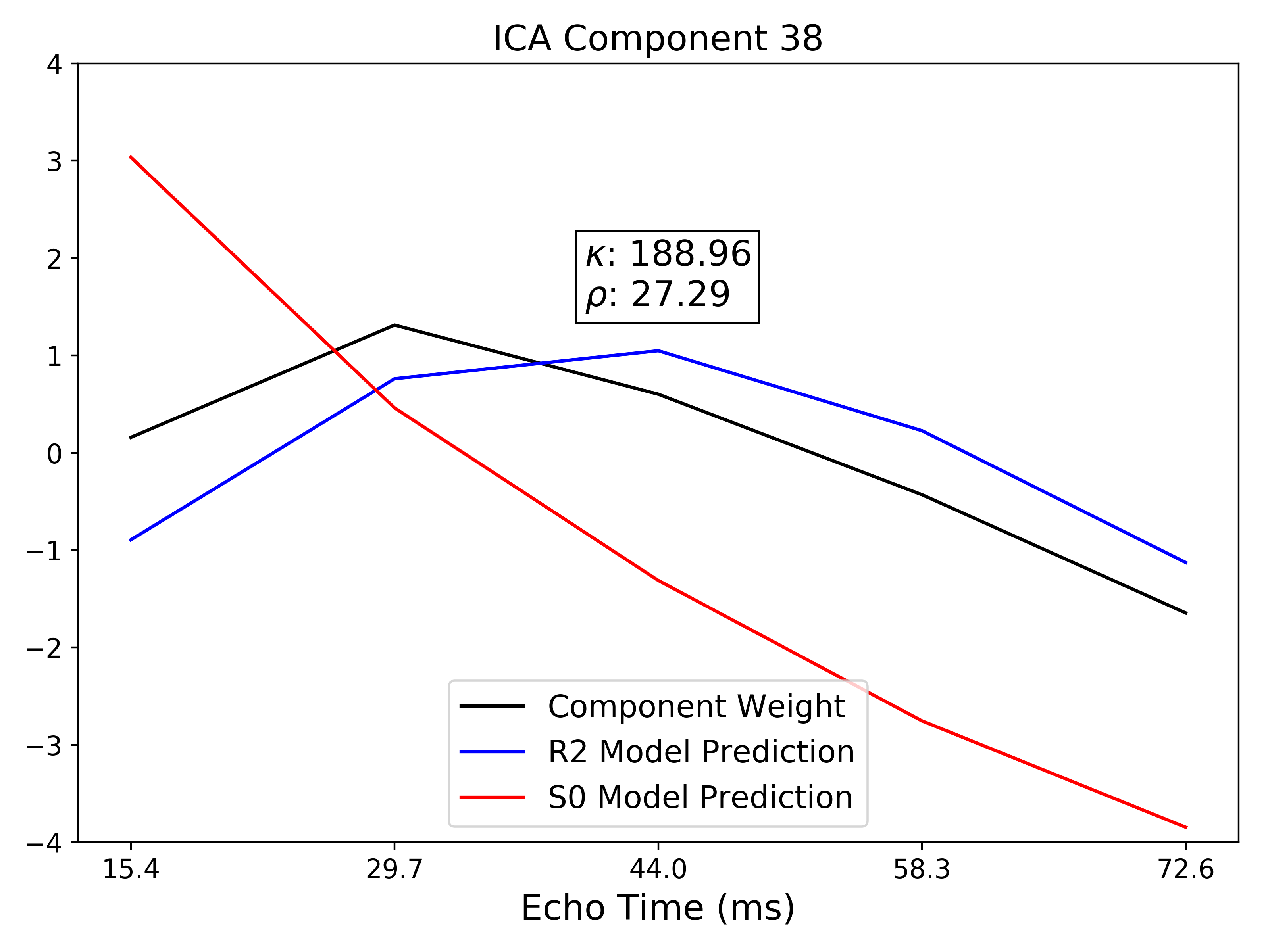
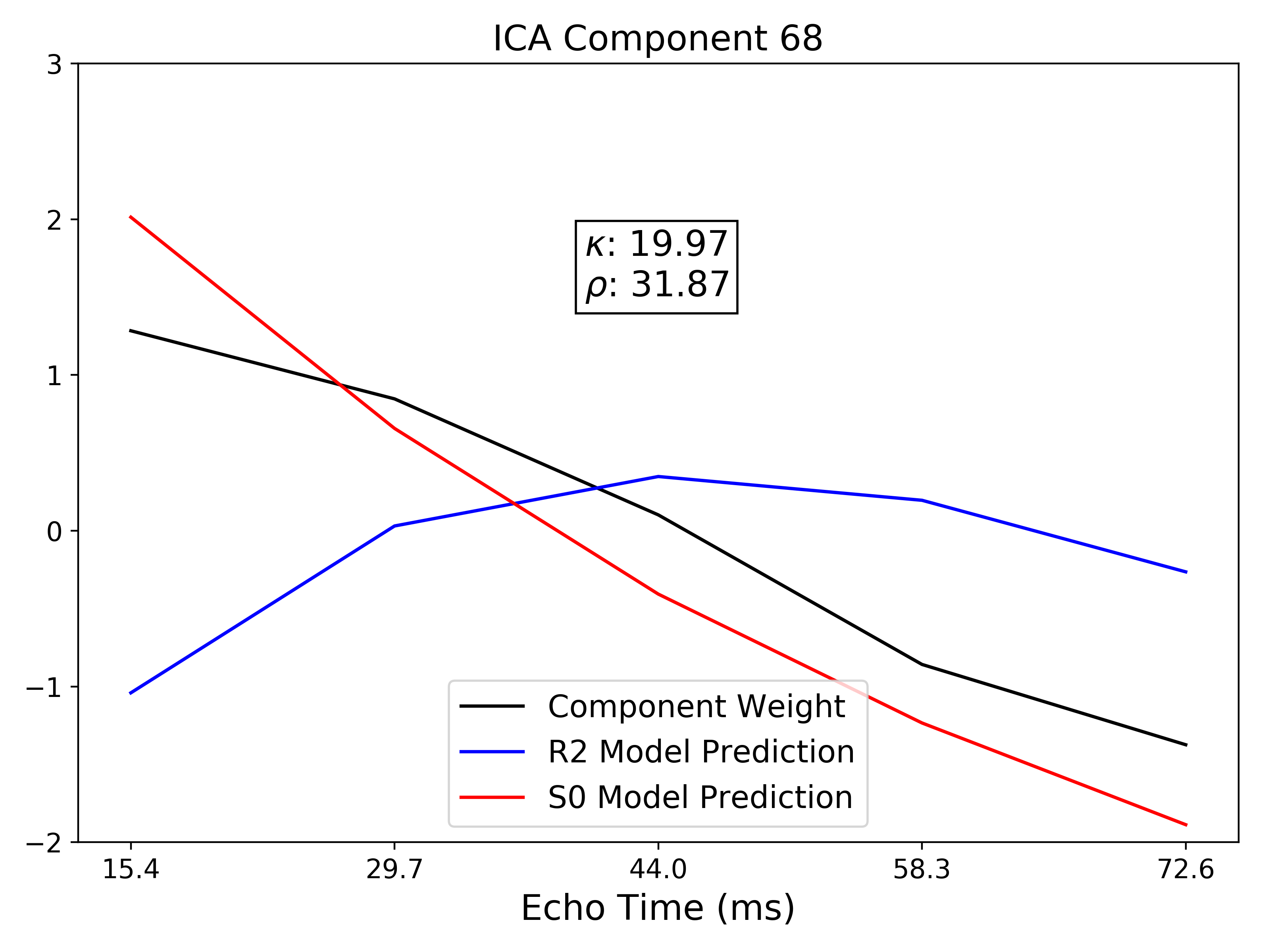
A decision tree is applied to  ,
, 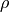 , and other metrics in order to
classify ICA components as TE-dependent (BOLD signal), TE-independent
(non-BOLD noise), or neither (to be ignored). The actual decision tree is
dependent on the component selection algorithm employed.
, and other metrics in order to
classify ICA components as TE-dependent (BOLD signal), TE-independent
(non-BOLD noise), or neither (to be ignored). The actual decision tree is
dependent on the component selection algorithm employed. tedana includes
two options: kundu_v2_5 (which uses hardcoded thresholds applied to each of
the metrics) and kundu_v3_2 (which trains a classifier to select components).
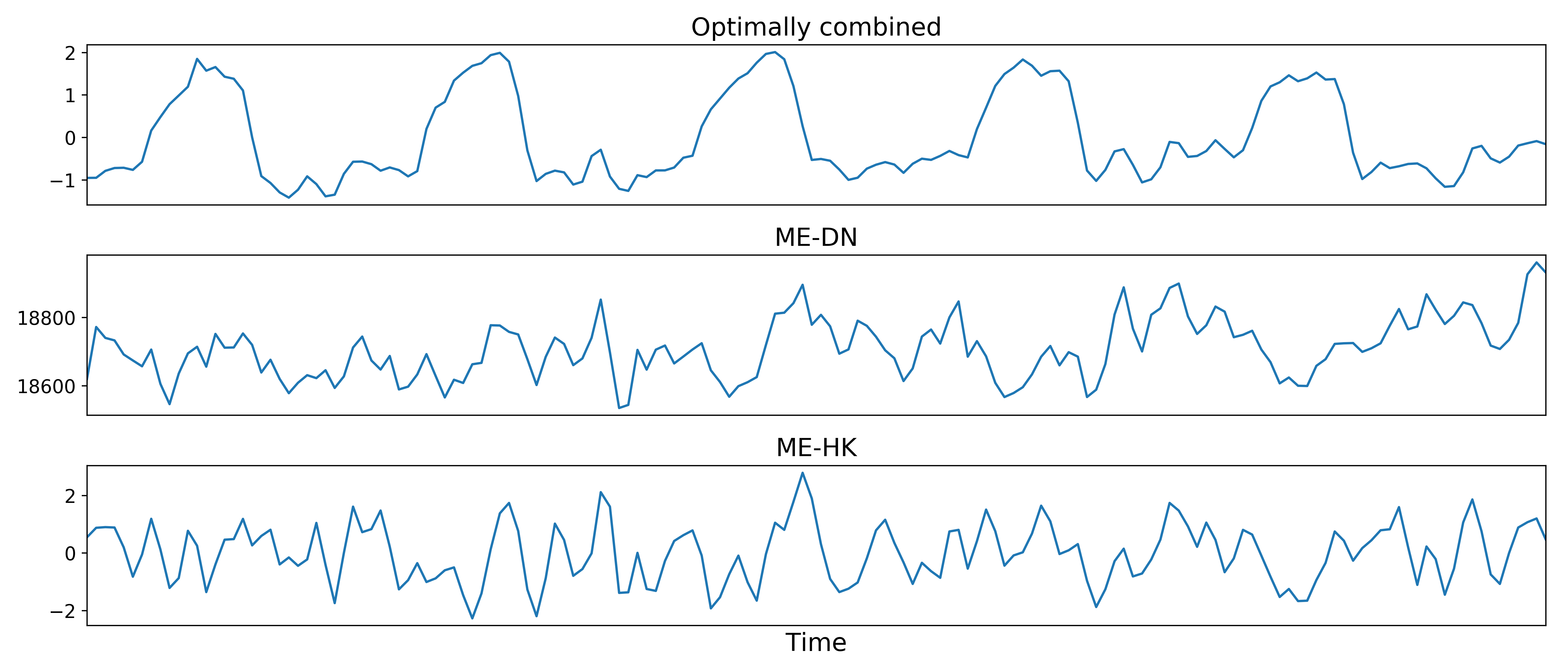
Removal of spatially diffuse noise (optional)¶
Due to the constraints of ICA, MEICA is able to identify and remove spatially localized noise components, but it cannot identify components that are spread out throughout the whole brain. See Power et al. (2018) for more information about this issue. One of several post-processing strategies may be applied to the ME-DN or ME-HK datasets in order to remove spatially diffuse (ostensibly respiration-related) noise. Methods which have been employed in the past include global signal regression (GSR), T1c-GSR, anatomical CompCor, Go Decomposition (GODEC), and robust PCA.