tedana: TE Dependent ANAlysis¶
The tedana package is part of the ME-ICA pipeline, performing TE-dependent
analysis of multi-echo functional magnetic resonance imaging (fMRI) data.
TE-dependent analysis (tedana) is a Python module for denoising
multi-echo functional magnetic resonance imaging (fMRI) data.



About¶

tedana originally came about as a part of the ME-ICA pipeline.
The ME-ICA pipeline originally performed both pre-processing and TE-dependent
analysis of multi-echo fMRI data; however, tedana now assumes that you’re
working with data which has been previously preprocessed.
Citations¶
When using tedana, please include the following citations:
tedana Available from: https://doi.org/10.5281/zenodo.1250561
![]()
2. Kundu, P., Inati, S. J., Evans, J. W., Luh, W. M. & Bandettini, P. A. (2011). Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. NeuroImage, 60, 1759-1770.
3. Kundu, P., Brenowitz, N. D., Voon, V., Worbe, Y., Vértes, P. E., Inati, S. J., Saad, Z. S., Bandettini, P. A., & Bullmore, E. T. (2013). Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proceedings of the National Academy of Sciences, 110, 16187-16192.
Alternatively, you can automatically compile relevant citations by running your
tedana code with duecredit. For example, if you plan to run a script using
tedana (in this case, tedana_script.py):
python -m duecredit tedana_script.py
You can also learn more about why citing software is important.
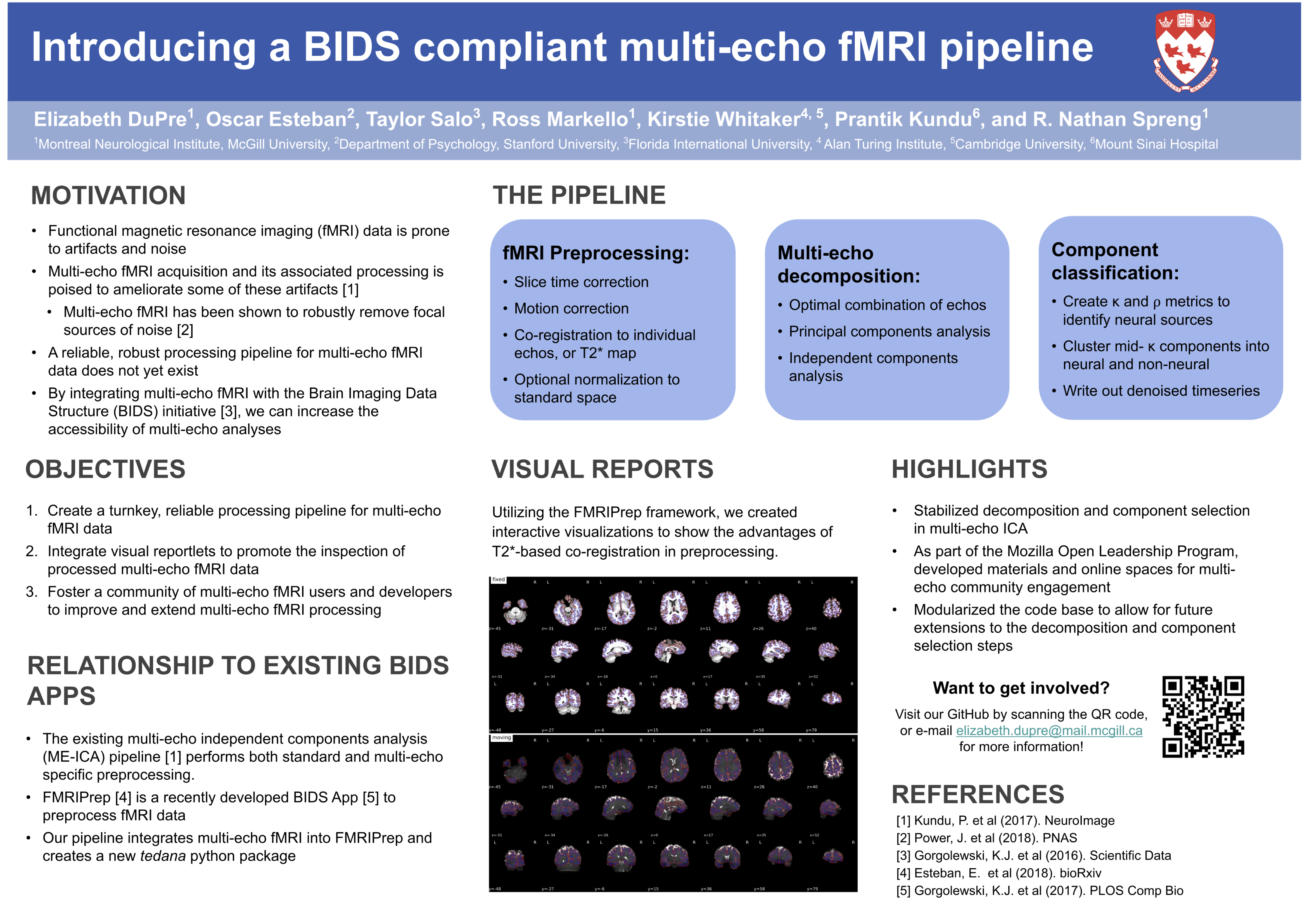
Posters¶
License Information¶
tedana is licensed under GNU Lesser General Public License version 2.1.